As clinical SAS programmer, we sometimes need to import and parse annotations contained in the Annotated Case Report Form (aCRF) for creating or validating Define.xml. When parsing the imported comments from aCRF, our ultimate goal is to identify the variable. Then we can get the correspondence information, such as CRF page. In this post, I’ll introduce a method using Perl regular expression. Syntax: PRXCHANGE (regular-expression-id|perl-regular-expression, times, source). Example:
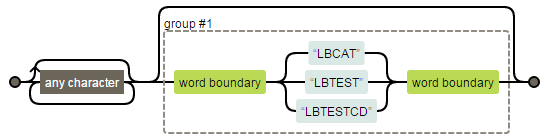
COMMENTS=prxchange("s/.*?(\b(?:LBCAT|LBTEST|LBTESTCD)\b)?/\1 /o", -1, cats(COMMENTS));Regular expression visualization by Regexper:
Here’s a brief explanation of the regular expression used in the example above. The “.” matches any single character except newline. The “*?” is lazy repetition factor, matches 0 or more occurrences of the preceding character as few times as possible. The first “(“and “)” characters matches a pattern and creates a capture buffer for the match. The last “?” is greedy repetition factor, matches the first capturing group zero or one time as many times as possible. The “\b” matches a word boundary. Since we want to mention “\b” only once, so the second “(“and “)”characters are needed. The “(?:…)” means non-capturing group, the “?:” is not necessary in this example. Since there is no memory required for the second catch (?:), it may work faster. The “\1” matches capture buffer 1.